BioInfToolServer
KIPEs
KIPEs (Knowledge-based Identification of Pathway Enzymes) enables the automatic identification and annotation of flavonoid biosynthesis genes/proteins. Users only need to supply a FASTA file with the peptide or transcript sequences. Please read the original publication for details: Pucker et al., 2020. KIPEs is also available for download from a GitHub repository.
Example dataset:
MYB_annotator
Our automatic MYB annotation workflow allows users to supply a FASTA file of peptide or transcript sequences to identify the MYB transcription factors. Please have a look at the original publication for details: Pucker, 2022. MYB_annotator is also available for download from a GitHub repository.
Example dataset:
CoExp
CoExp enables users to identify co-expressed genes. A list of genes can be provided and the analysis is performed for each of them. All genes with a sufficient co-expression with the candidate gene are returned. A functional annotation file can be provided to assign a functional annotation to the results. CoExp is also available for download from a GitHub repository.
Example dataset:
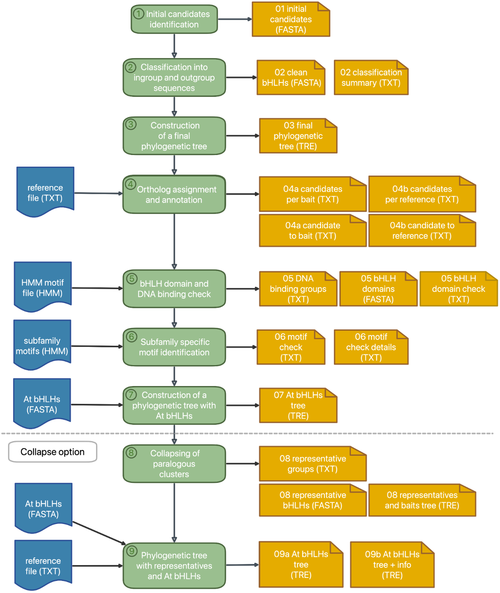
bHLH_annotator
Our automatic bHLH annotation workflow allows users to supply a FASTA file of peptide or transcript sequences to identify the bHLH transcription factors. Please have a look at the original publication for details: Thoben & Pucker, 2023. bHLH_annotator is also available for download from a GitHub repository.
Example dataset:
NAVIP
NAVIP (Neighborhood-Aware Variant Impact Predictor) processes a variant calling format (VCF) file and predicts the functional impact of the sequence variants on annotated genes. Additional details can be found in the original publication: Baasner et al., 2019. NAVIP is also available for download from a GitHub repository.
Example dataset:
Feel free to contact us via email if you have any questions or comments related to our tools. Are you missing a feature or one of our tools? Please get in touch! We are planning to extend the collection of offered tools. Learning about the interest in different tools helps us to prioritize the addition of new tools.

